Project 1: Protein Engineering and Development of Antigen Specific Therapeutics and Novel Therapeutic Approaches of Autoimmune Disorders
Autoimmune disease affects at least 23.5 million Americans, and is the 2nd leading cause of long-term chronic illness. Autoimmune disease is caused when a person’s immune system begins to attack healthy cells in the body. Current therapies for autoimmune disease often utilize non-specific drugs that suppress the entire patient’s immune system. This approach increases the risk of patients contracting other infections more easily and decreases their ability to fight infections off successfully. Thus, the goal of our research is to develop more targeted therapeutic approaches that will only suppress the aberrant portion of a patient’s immune system. In this project we focus on engineering proteins that can serve as diagnostic agents, helps us understand the function of relevant proteins involved in an autoimmune disorder, or that can inhibit the immune system from attacking healthy cells. The MIR lab currently focuses on developing therapeutics for primary membranous nephropathy and systemic lupus erythematosus.
Collaborators:
Laurence Beck, Jr.: Boston University Medical College
Roberto de la Salud Bea: Rhodes College
Kim Brien: Rhodes College
Gérard Lambeau: Institut de Pharmacologie Moléculaire et Cellulaire (France)
Project 2: Modelling and Investigation of Structure and Function Relationships of Unsolved Protein Structures
Understanding protein structures associated with autoimmune disorders, cancer, and neurological disorders is essential to developing better therapeutics. Many of the proteins of interest in these disorders are uncharacterized. Thus, my lab also focuses on developing 3D-models of protein structures related to cancer and protein antigens involved in autoimmune disorders. Recently, we successfully developed a homology model of the 250kDa thrombospondin type-1 containing domain 7A antigen involved in the autoimmune disorders idiopathic membranous nephropathy (IMN). Currently we are focused on using these models to understand the biological function of these proteins and other similar proteins in this super protein family.
Collaborators:
Laurence Beck, Jr.: Boston University Medical College
Publications from this project:
Shana V. Stoddard, Colin L. Welsh, Maggie M. Palopoli, Serena D. Stoddard, Mounika P. Aramandla, Riya M. Patel, and Laurence H. Beck, Jr.Structure and Function Insights Garnered from In silico Modelling of the Thrombospondin Type-1 Domain-Containing 7A Antigen. PROTEINS: Structure, Function, Bioinformatics. Proteins. 2018 Dec 06. PMID: 3052053
Project 3: Design, Synthesis, and Biological Testing of Novel Therapeutic Compounds for Immune-Mediated Diseases and Neurological Disorders.
Cancer is the second leading cause of death in the United States. In the MIR lab we develop compounds that could be used to treat several cancer types. Our compound are designed to target a family of 18 enzymes called histone deacetylases (HDAC) that are important for gene regulation. It has been shown that different types of cancers vary in the amount and type of HDAC enzymes they express, making them good drug targets for cancer therapy. Unfortunately there have been challenges in designing HDAC inhibitors that can target only one specific HDAC enzyme out of the family of 18. Thus, this project focuses on development of not only potent HDAC inhibitors but also selective HDAC inhibitors. One of the specific cancers that the MIR lab targets is neuroblastoma which, has been shown to have over expression of HDAC8. Neuroblastoma is a childhood pediatric cancer, which has proved difficult to treat. Therefore, we are developing selective HDAC8 inhibitors.
Neurological disorder are also impacted by the histone deacetylase family of enzymes. In particular histone deacetylase 4 (HDAC4) has been found to be a relevant target for Huntington’s disorder and glioblastoma a cancer of the nervous system. Recently we have identified a series of novel HDAC4 inhibitor compounds and produced guidelines for better HDAC4-selective inhibitors. Currently the MIR lab is focused on synthesis and biological evaluation of our designed HDAC4-selective inhibitors that would be ideal for the neurological disorder Huntington’s disease and glioma tumors.
Collaborators:
Davita Watkins: University of Mississippi
Roberto de la Salud Bea: Rhodes College
Publications from this project:
Sivaraman Balasubramaniam, Sajith Vijayan, Liam Goldman, Xavier A. May, Kyra Dodson, Sweta Adhikari, Davita L. Watkins, and Shana V. Stoddard. Design and Synthesis of Diazine-based Panobinostat Analogues for HDAC8 Inhibition. Belstein Journal of Organic Chemistry, 2020, 16(1), 628-637
In silico Design of Novel Histone Deacetylase 4 Inhibitors: Design Guidelines for Improved Binding Affinity. Shana V. Stoddard, Kyra Dodson, Kamesha Adams, and Davita L. Watkins. International Journal of Molecular Science, 2020 21(1), 219
Shana V. Stoddard, Xavier A. May, Fatima Rivas, Kyra Dodson, Sajith Vijayan, Swetha Adhika, Kordarius Parker, Davita L. Watkins. Design of Potent Panobinostat Histone Deacetylase Inhibitor Derivatives: Molecular Considerations for Enhanced Isozyme Selectivity between Histone Deacetylase 2 (HDAC2) and Histone Deacetylase 8 (HDAC8) Molecular Informatics, 2018 38(3), 1800080
Project 4: Development of Broad-Spectrum Coronavirus Antivirals

Coronaviruses have caused one epidemics and two pandemics in the last 20 years. The COVID-19 pandemic has by far been the most significant threat to the way of life of billions across the globe. To address the need for a therapeutic option work in the MIR lab is defining the substrate preferences of the coronavirus main proteases (Mpro). We are developing novel antivirals that target the main protease using computational chemistry, synthesis, and biological evaluation. We are also pursuing development of novel proteins which bind to the coronavirus crown to prevent the crown protein from being able to invade cells.
Collaborators:
Davita Watkins: University of Mississippi
Sudeshna Roy: University of Mississippi
Publications from this project:
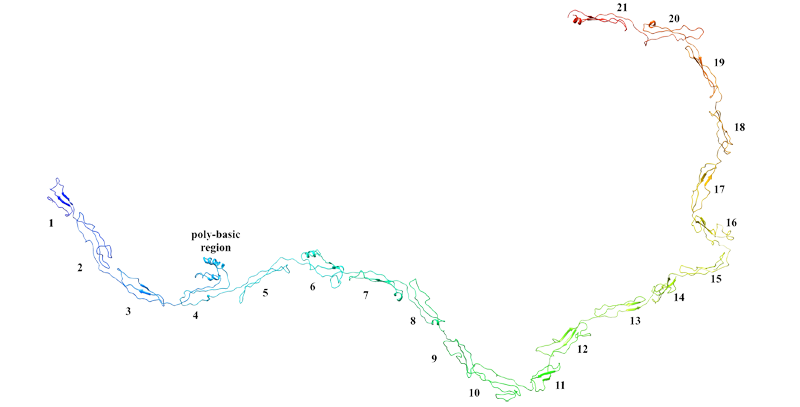
Shana V. Stoddard, Felissa E. Wallace*, Serena D. Stoddard, Qianyi Cheng, Daniel Acosta, Shaliz Barzani, Marissa Bobay, Jared Briant, Christian Cisneros, Samantha Feinstein, Kelsey Glasper, Munazza Hussain, Abigail Lidoski, Pranay Lingareddy, Grace Lovett, Leslie Matherne, Jackson McIntosh, Nikita Moosani, Lia Nagge, Kudzai Nyamkondiwa, Isaiah Pratt, Emma Root, Mary Rose Rutledge, Mackenzie Sawyer, Yash Singh, Kristiana Smith, Ubaid Tanveer, and Sona Vaghela. In silico design of peptide-based SARS-CoV-2 fusion inhibitors that target WT and mutant versions of SARS-CoV-2 HR1 domains. Biophysica, 2021, 1(3), 311-327
Shana V. Stoddard, Serena D. Stoddard, Benjamin K. Oelkers, Kennedi Fitts, Kellen Whalum, Kaylah Whalum, Alexander D. Hemphill, Jithin Manikonda, Linda Michelle Martinez, Elizabeth G. Riley, Caroline M. Roof, Nowreen Sarwar, Doni M. Thomas, Emily Ulmer, Felissa E. Wallace*, Pankaj Pandey, and Sudeshna Roy. Optimization Rules for SARS-CoV-2 Mpro Antivirals: Ensemble Docking and Exploration of the Coronavirus Protease Active Site. Viruses, 2020, 12, 942